General Theory of Life
Three cornerstones of life

Albert Szent-Györgyi
MOLECULAR DYNAMIC OF BIOLOGICAL MACROMOLECULES
- Biological macromolecules (proteins, DNA) are electron conductors.
- The movement of delocalized electrons through the conducting band of biological macromolecules causes its dynamic.
- Molecular dynamics is a basic prerequisite for communication between biological macromolecules

Herbert Fröhlich
SELECTIVE LONG-DISTANCE INTERACTION BETWEEN BIOLOGICAL MOLECAL
- The dynamic of biological molecule transfers to the water and propagates as a dipole oscillation on the long distance around the molecule.
- Between biological molecules oscillating with the same frequency the attractive forces are generating (the resonant interaction).
- The resonant interaction allows highly specific and effective recognition and targeting between molecules in the biological space overcrowded with billons of molecules.

Hermann Emil Fischer
SPECIFIC CHEMICAL BINDING BETWEEN BIOLOGICAL MOLECULES
- The specific chemical binding between biological molecules is determined by their structural complementarity (the lock and key concept).
The legacy of Albert Szent-Györgyi
I expect that the next great stride of biology will be its shift to the electronic dimension. This will be the step by which biology truly enters the realm of modern medicine. Probably this Electronic biology will be the biology of the 21st century.
– Albert Szent-Györgyi, 1969
An electron, given off easily, has a high energy level, while an orbital which tends to take up an electron, has a low energy level. The transmitted electron thus goes from a high to a low level, and releases the energy which corresponds to the difference of the two levels. It is this energy which drives life.
– Albert Szent-Györgyi, Electronic Biology and Cancer, Marcel Dekker Inc. 1976

The main postulates of the electronic biology determined by Albert Szent-Györgyi
- Biological molecules (proteins, DNA and RNA) are electron conductors
- The function of biological molecules is determined by their electronic properties
- Electronic properties/function of biological molecules are modulated by electron acceptors and electron donors
- The first proteins which formed on the Earth were weak electron conductors with the saturated conduction band
- The oxygen was the first electron acceptor which increased the electronic conductivity of proteins by desaturation of their conduction band
- The increased electron conductivity of proteins allowed the greater number of their interactions with other molecules, which was the main prerequisite for the emergence of life on the Earth
- Proteins that are weak electron conductors with the saturated conduction band are the hallmark of cancer
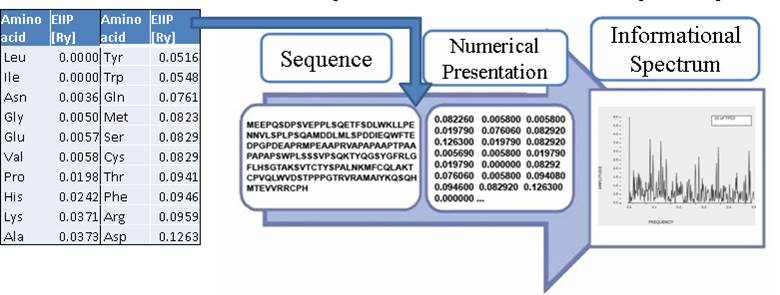
Electronic molecular descriptors
The electron-ion interaction is the basis for almost all processes in condensed mater. For this reason, the knowledge of the electron-ion interaction potential (EIIP) is one of the main problems of the condensed-matter physics. Due to the EIIP complexity, this problem has not been solved on the grounds of the first principles. The lack of the basic theory has been compensated by the development of several phenomenological potentials.
The General Model Pseudopotential (GMP) is the simplest phenomenological potential proposed in 1972 by Veljkovic [1,2]:
W = 0.25Z*sin(1.04ϖZ*)/2ϖ (1)
where Z* is the average quasivalence number (AQVN) determined as
Z* = ΣmniZi/N (2)
where Zi is the valence number of the i-th atomic component, ni is the number of atoms of the i-th component, m is the number of atomic components in the molecule, and N is the total number of atoms. The EIIP values calculated according to equations (1) and (2) are in Rydbergs (Ry).
The GMP was successfully used in the study of different complex phenomena in solid state physics and material science (e.g. phonon spectra, superconductivity, electronic properties of metals, nuclear waste management, etc.).
The well-known fact from the theory of condensed matter that all phenomena are basically dependent upon the characteristic of EIIP led to the assumption that also in biological processes this physical interaction plays an important role [3]. Based on this assumption, the number of delocalized electrons, represented by AQVN, and their energy, represented by EIIP, were proposed as the fundamental molecular descriptors of biological molecules [3].
References
- Veljkovic V, Slavic I (1972) Simple general-model pseudopotential. Phys Rev Lett 29: 105-106.
- Veljkovic V (1973) The dependence of the Fermi energy on the atomic number. Phys Lett 45A: 41-42.
- Veljkovic V (1980) Theoretical approach to preselection of cancerogens and chemical carcinogenesis. Gordon & Breach, New York.
More Information about Molecular Descriptors
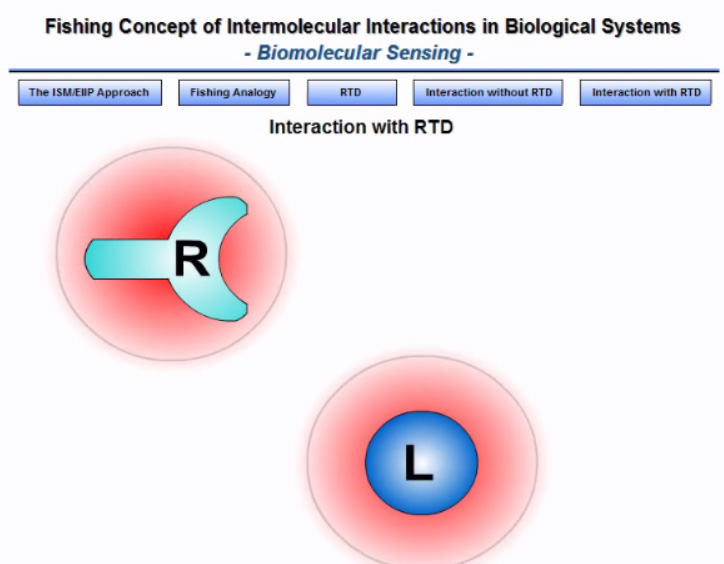
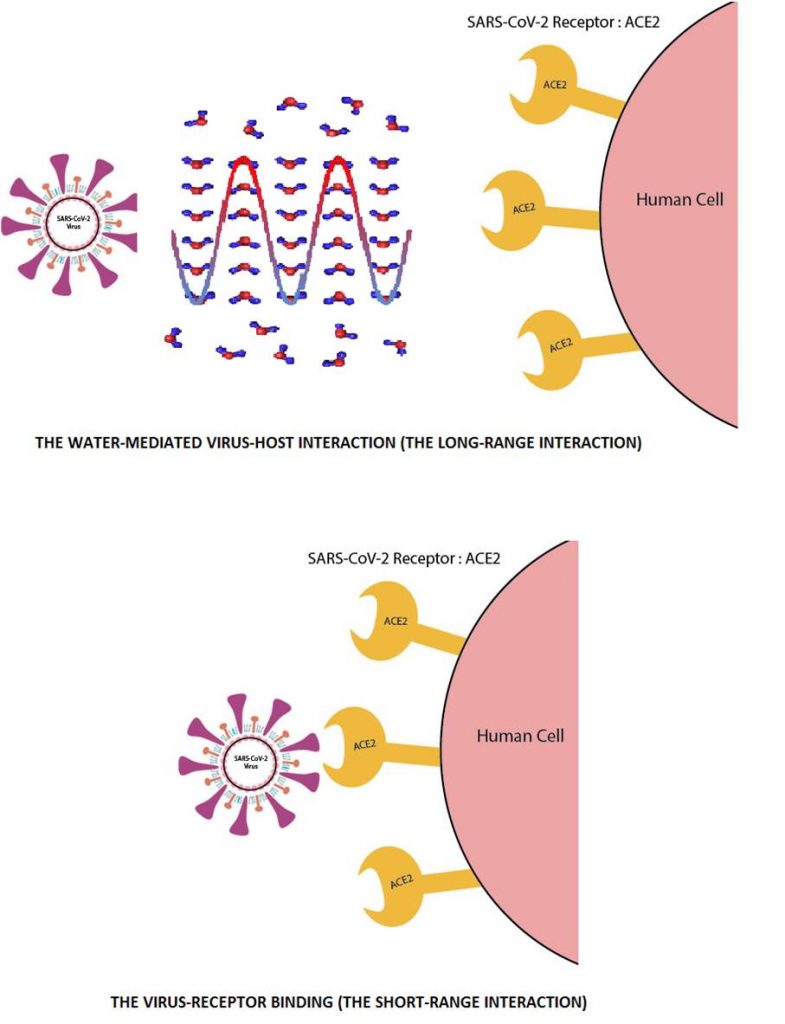
The long-distance molecular interactions
Getting to the root of the problem
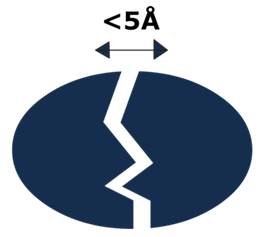
According to the current concept, the short-range electrostatic forces (distances <5Å) are the key factors that determine the interaction between biological molecules.
A solution of the problem
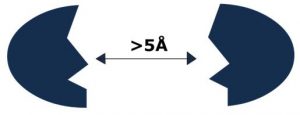
Physicist Frölich proposed an acceptable solution to this problem. He suggested the resonant interaction between biological molecules, which lead to the appearance of frequency-selective long-range attractive forces that are efficient at a distance longer than one linear dimension of the interacting macromolecules (100 – 1000 Å). These forces allow an increase in the number of productive collisions in comparison with the accidental encounter of molecules.
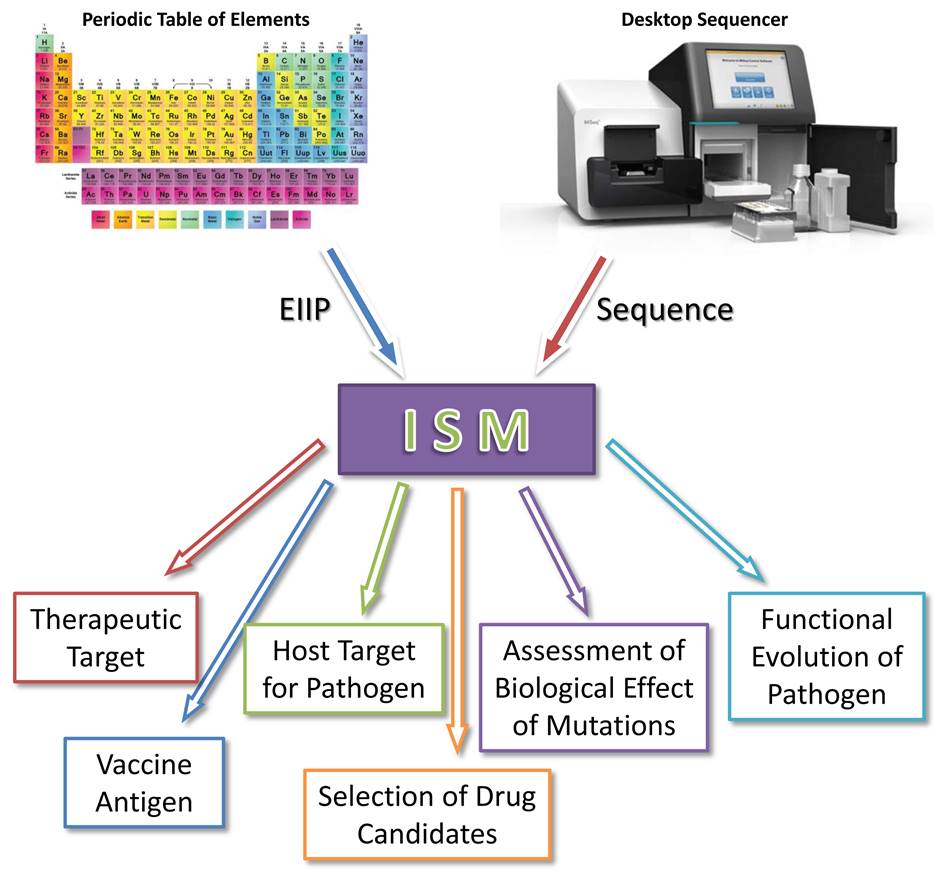
The electronic biology-based tool for study of biological molecules
[Veljkovic V., Cosic I., Dimitrijevic B., Lalovic D., Is it possible to analyze DNA and protein Sequence by the method of digital signal processing?. IEEE Trans., BME-32, 337 (1985).]
Advantages of the electronic biology concept in the prevention and therapy of chronic and infectious diseases
Development of the new generation of drugs and vaccines that block the recognition and targeting between proteins
Conventional concept og the drug development
- Chronic diseases: Block ligand – therapeutic target binding
- Infectious diseases: Blok infectious agent – host proteins binding

Novel concept of the drug design
- Chronic diseases: Block ligand – therapeutic target recognition and targeting
- Infectious diseases: Blok infectious agent – host proteins recognition and targeting
The drugs that block the long-range molecular interactions are less sensitive to mutations compared to the drugs that block molecular binding.
Minimal input data (only sequences and the atomic numbers) allows characterization of novel pathogens, monitoring of their functional evolution and determination of their therapeutic and vaccine target without any prior experimental knowledge.
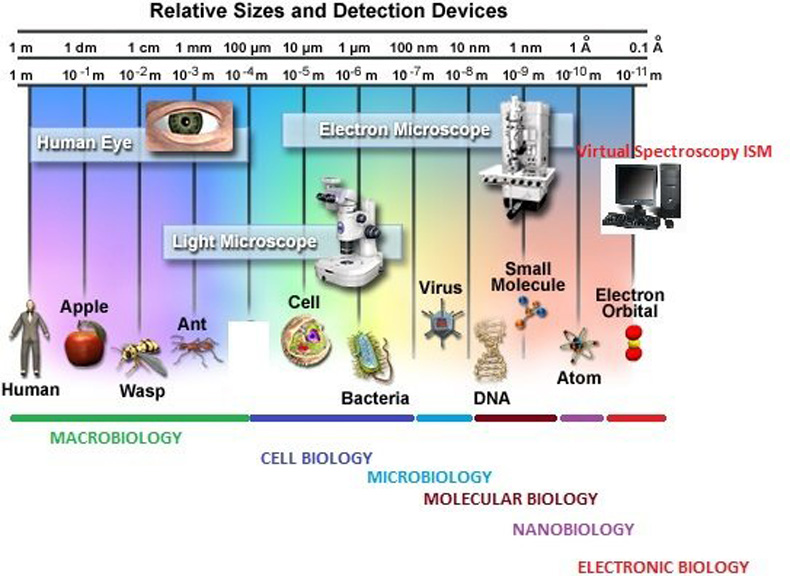
Relative Sizes and Scientific Fields